Part:BBa_K1033002
stilbene synthase (STS) with RBS
This part codes for the enzyme stilbene synthase (STS). Stilbene synthase is the third and last enzyme in the stilbene synthesis pathway which derrives from the universal polyphenol biosynthetic pathway. It catalyzes the formation of resveratrol, using the substrates coumaryl-CoA and malonyl-CoA. This version is derived from vitis vinifiera, or normally called red grape[1]
In our project, we have been using it together with 4-coumarate ligase, that produces coumaryl-CoA. From this metabolite, stilbene synthase can produce resveratrol.
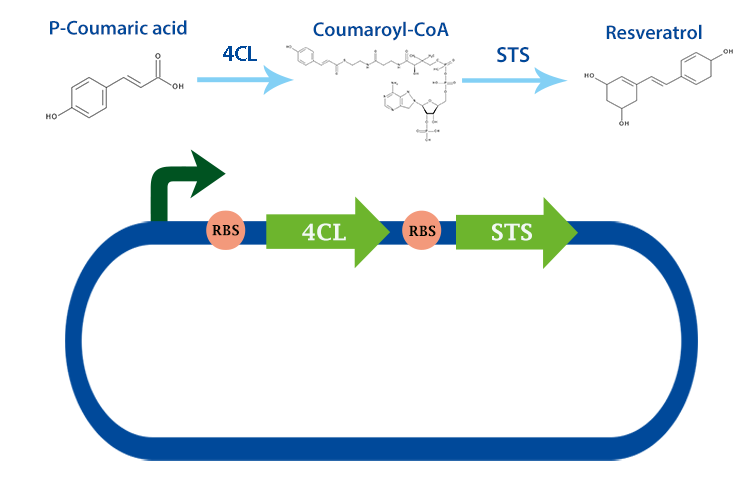
Applications With the help of this enzyme one can produce the healthy antioxidant resveratrol derived from redwhine. For the production of resveratrol, coumaryl-CoA can be obtained from our other biobrick, 4Cl. [2]
An improved version if this part is reported on the page: https://parts.igem.org/Part:BBa_K4388003
KCL iGEM 2022
The stilbene synthase (STS) from Vitis vinifera has been extensively characterised for its production of resveratrol within resveratrol- or pterostilbene-yielding biosynthetic pathways in engineered microorganisms (Feng et al., 2022). Comparisons with stilbene synthases from other species, including the also well-characterised Arachis hypogaea STS, have demonstrated a greater resveratrol titre from VvSTS expression (Lim et al., 2011; Camacho-Zaragoza et al., 2016). With the VvSTS family having already been largely structurally and functionally characterised (Parage et al., 2012), more recent studies have investigated further improvements of VvSTS kinetics through mutagenesis approaches (Bhan et al., 2015a, 2015b; Yan et al., 2021).
Bhan et al. (2015a, 2015b) employed rational redesign strategies to expand both the cyclisation as well as the substrate binding pocket, using site-directed mutagenesis. However, these demonstrated decreased resveratrol synthesis while increasing the production of other compounds. On the other hand, Yan et al. (2021) recently generated a T50I/V170A VvSTS mutant (T51I/V171A in this BioBrick part due to an extra methionine at amino acid position 50) via random mutagenesis, increasing the yield of resveratrol 2-fold compared to the wild type (Yan et al., 2021). This was suggested to be due to the V170A mutation’s position at the bottom of the active site, with the decreased residue size expanding the active site for substrate binding.
Subcellular Localisation VvSTS
Similarly we used the same general-to-specific approach while using computational prediction approaches to confidently characterize another on of the enzymes
Citations
Feng, C., Chen, J., Ye, W., Liao, K., Wang, Z., Song, X., & Qiao, M. (2022). Synthetic Biology-Driven Microbial Production of Resveratrol: Advances and Perspectives. Frontiers in bioengineering and biotechnology, 10, 833920. https://doi.org/10.3389/fbioe.2022.833920
Lim, C. G., Fowler, Z. L., Hueller, T., Schaffer, S., & Koffas, M. A. (2011). High-yield resveratrol production in engineered Escherichia coli. Applied and environmental microbiology, 77(10), 3451–3460. https://doi.org/10.1128/AEM.02186-10
Camacho-Zaragoza, J. M., Hernández-Chávez, G., Moreno-Avitia, F., Ramírez-Iñiguez, R., Martínez, A., Bolívar, F., & Gosset, G. (2016). Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microbial cell factories, 15(1), 163. https://doi.org/10.1186/s12934-016-0562-z
Parage, C., Tavares, R., Réty, S., Baltenweck-Guyot, R., Poutaraud, A., Renault, L., Heintz, D., Lugan, R., Marais, G. A., Aubourg, S., & Hugueney, P. (2012). Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant physiology, 160(3), 1407–1419. https://doi.org/10.1104/pp.112.202705
Bhan, N., Li, L., Cai, C., Xu, P., Linhardt, R. J., & Koffas, M. A. (2015). Enzymatic formation of a resorcylic acid by creating a structure-guided single-point mutation in stilbene synthase. Protein science : a publication of the Protein Society, 24(2), 167–173. https://doi.org/10.1002/pro.2600
Bhan, N., Cress, B. F., Linhardt, R. J., & Koffas, M. (2015). Expanding the chemical space of polyketides through structure-guided mutagenesis of Vitis vinifera stilbene synthase. Biochimie, 115, 136–143. https://doi.org/10.1016/j.biochi.2015.05.019
Yan, Z. B., Liang, J. L., Niu, F. X., Shen, Y. P., & Liu, J. Z. (2021). Enhanced Production of Pterostilbene in Escherichia coli Through Directed Evolution and Host Strain Engineering. Frontiers in microbiology, 12, 710405. https://doi.org/10.3389/fmicb.2021.710405
Characterizaion data
Summary This enzyme was cloned out from a template with pcr provided from conrado.et.al[3]
Although we managed to clone out and sequence verify the genes for resveratrol production, we have had some problems in the characterization. The results are unclear, and we did not have time for further investigations.
Biobrick
We managed to biobrick and sequence verify this part.
Western blot
We also succeeded in expressing the enzyme stilbene synthase in E. coli. Our expression of the protein was however very weak, and due to time constraints we were not able to optimize our experiment.
To enable the detection of this protein by anti-his antibodies, 6-histidine tags was incorporated in the sequence.
We expressed our protein with a promotor working in both lactobacillus and e-coli. This way, you can easily transfer stilbene synthase to lactobacillus later on.
The size of our protein was calculated using ProtParam [5], 43 kDA.

Figure 1:Number 2 shows a very weak band of our protein at around 43 kDA. Positive control -> 1, Stilbene synthase -> 2
High pressure liquid chromatography
We tested our biobrick 4Cl-STS on HPLC, by adding p-coumaric acid as a precursor. The result we saw was quite unclear. We saw that the e-coli produced something out of the ordinary, but the absorbation was low and the peaks did not exactly match the standard. The peak at around ~33 min could correspond to our standard, but it is unclear. We theorize it is something in the actual hplc measurement that fails, or that something happens to our resveratrol metabolite in our e-coli. This result could correspond to the poor results in our blot. We hope that iGEM teams can continue to work on these biobricks in the future.

Figure 2: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.

Figure 3: Resveratrol standard, peaks around ~33, ~34 min.
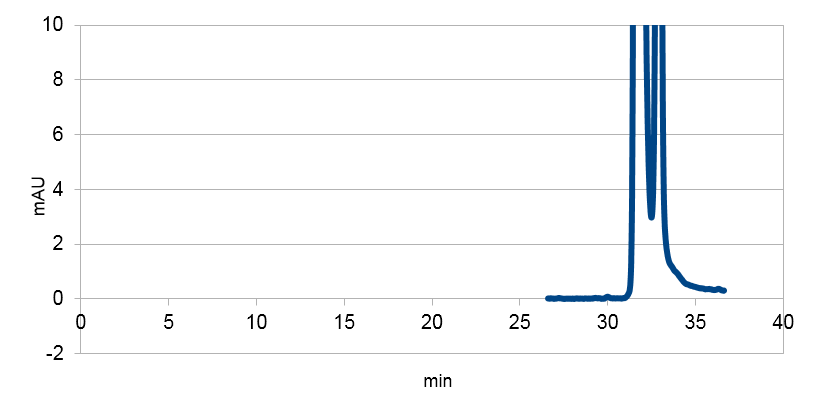
Figure 4: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.
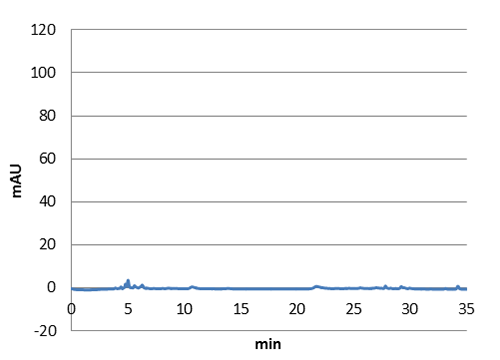
Figure 5: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes.
References
[1] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol: the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5
[2] Baur, Y, A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature Vol 444| 16 November 2006
[3] Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//collections/probiotics/production
| None |

